#install.packages(c("rpact", "doSNOW", "doRNG", "foreach"))
library(rpact)
library(doRNG)
reproducibleSimulationExample <- function(iterations = 100, seed = 4357829) {
cores <- parallel::detectCores(logical = FALSE)
parallelComputingCluster <- parallel::makeCluster(cores)
on.exit(parallel::stopCluster(parallelComputingCluster), add = TRUE)
doSNOW::registerDoSNOW(parallelComputingCluster)
alternatives <- seq(0.1, 0.8, 0.1)
set.seed(seed)
resultLists <- foreach::foreach(
index = 1:length(alternatives),
.verbose = FALSE, .packages = c("rpact"),
.export = c("getSimulationMeans")
) %dorng% {
alternative <- alternatives[index]
simulatedDesign <- getSimulationMeans(
alternative = alternative,
thetaH0 = 0,
plannedSubjects = 100,
maxNumberOfIterations = iterations)
data.frame(
alternative = alternative,
power = round(sum(simulatedDesign$overallReject), 5)
)
}
results <- c()
for (result in resultLists) {
results <- rbind(results, result)
}
results
}Parallelized Simulations with rpact
Gernot Wassmer
April 26, 2024
Introduction
- Project: A Phase III double-blind, randomized, placebo controlled, multi-center trial with adaptive sample size recalculation.
- Objective: To Evaluate the Efficacy and Safety of a New Therapy with an exponentially distributed endpoint through the usage of the Wilcoxon test.
- Key Performance Metrics and Requirements from FDA:
- Achieved power.
- Expected sample sizes and stopping probabilities.
- Confidence interval coverage probabilities and bias of estimates.
- Type I error rate control.
- Simulation with R through utilization of
rpact.
Simulation with R
- Function Developed:
getSimulateAdaptiveDesign(). - Purpose: To simulate test characteristics of the adaptive design with data-driven sample size recalculation.
- Comparison: Proposed design with a design with and without sample size reassessment
- Initial Simulation: 10,000 iterations took around 12 hours
- range of situations to be considered
- raw data generation
wilcox.test(),uniroot(), andgetFinalConfidenceInterval()requires capacity
Feedback and Challenges
- FDA Statistician’s Feedback: Perform the simulation again with 100,000 iterations within a week.
- Challenge: How to meet the new deadline with the increased computational demand?
Solution: Parallelization
- Approach: Utilization of R packages ‘parallel’, ‘doSNOW’, and ‘foreach’ for parallelization.
- Outcome: Enabled completion of the FDA-required simulation within the deadline.
- New Challenge: Ensuring reproducibility and seed safety of the simulations.
Final Resolution
- Solution: Utilization of the R package ‘doRNG’.
- Implementation: Replacing
%dopar%commands with%dorng%to ensure seed safety. - Result: Successful completion and reproducibility of the simulation, meeting FDA requirements.
Reproducible Simulation Example
alternative power
1 0.1 0.07331
2 0.2 0.16663
3 0.3 0.32539
4 0.4 0.51689
5 0.5 0.71478
6 0.6 0.84642
7 0.7 0.93811
8 0.8 0.98015Study Simulation Results


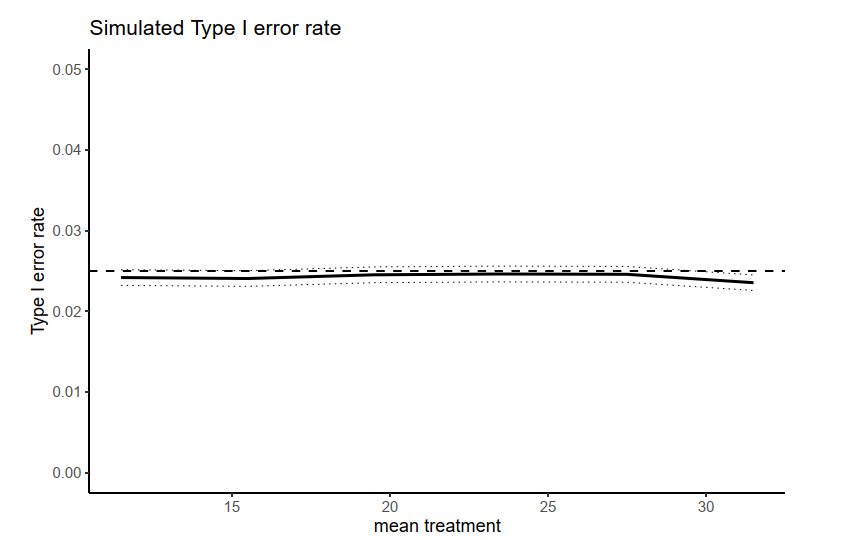
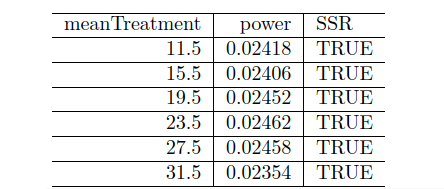
Goal Achieved

100,000 simulations took around 14 hours
Summary
rpactis a comprehensive validated package for designing group sequential designs, with a focus on adaptive extensions using the inverse normal method or Fisher’s combination test.- Usage supported by extensive documentation and vignettes, ongoing development.
- RPACT Cloud suitable for learning R commands and
rpactas a stand-alone. - Computation time can be substantially improved by parallel computing.
- Rich R extension tools for performing more advanced strategies (e.g., “reproducable results”, promizing zone approach, etc.).
- Comes along with the more and more accepted usage of R in pharmaceutical research and the transition towards R in many institutions.
